A community challenge for a pancancer drug mechanism of action inference from perturbational profile data
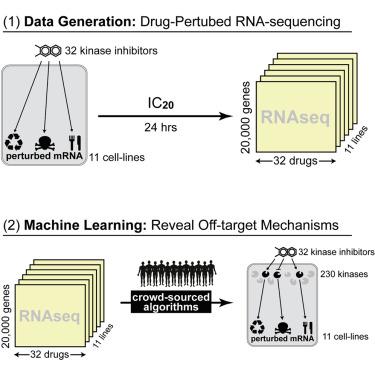
Douglass Jr. EF, Allaway RJ, Szalai B, Wang W, Allaway RJ, Szalai B, Wang W, Tian T, Fernández-Torras A, Realubit R, Karan C, Zheng S, Pessia A, Tanoli Z, Jafari M, Wan F, Li S, Xiong Y, Duran-Frigola M, Bertoni M, Badia-i-Mompel P, Mateo L, Guitart-Pla O, Chung V, DREAM CTD-squared Pancancer Drug Activity Challenge Consortium, Tang J, Zeng J, Aloy P, Saez-Rodriguez J, Guinney J, Gerhard DS, Califano A, The Columbia Cancer Target Discovery and Development (CTD2) Center is developing PANACEA, a resource comprising dose-responses and RNA sequencing (RNA-seq) profiles of 25 cell lines perturbed with ∼400 clinical oncology drugs, to study a tumor-specific drug mechanism of action. Here, this resource serves as the basis for a DREAM Challenge assessing the accuracy and sensitivity of computational algorithms for de novo drug polypharmacology predictions. Dose-response and perturbational profiles for 32 kinase inhibitors are provided to 21 teams who are blind to the identity of the compounds. The teams are asked to predict high-affinity binding targets of each compound among ∼1,300 targets cataloged in DrugBank. The best performing methods leverage gene expression profile similarity analysis as well as deep-learning methodologies trained on individual datasets. This study lays the foundation for future integrative analyses of pharmacogenomic data, reconciliation of polypharmacology effects in different tumor contexts, and insights into network-based assessments of drug mechanisms of action.
Cell Reports Medicine,
2022, 3
Pubmed: 35106508
Direct link: 10.1016/j.xcrm.2021.100492
